
| SMOS NetCDF Conversion | 
|
The NetCDF Export Command can be used to convert the data of
SMOS products in Earth Explorer file format to NetCDF while
preserving the original grid point structure of the data.
The conversion tool supports the following SMOS product types:
The target file format for the conversion process is NetCDF-4 (compressed). General information about the NetCDF-4 format can be found at UNIDATA NetCDF (Network Common Data Form)
The NetCDF Export dialog can be invoked within SNAP using the "Tools" menu.
The main parameters are available in the I/O Parameters tab shown below. Moving the mouse pointer over parameter elements yields further information.
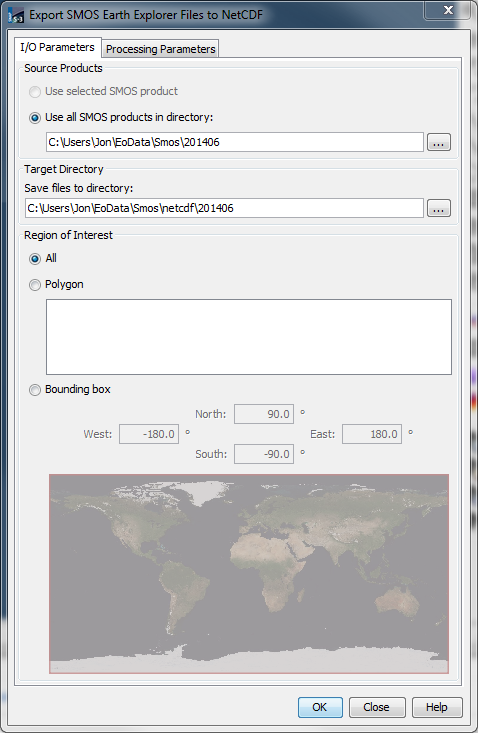
Source Products: Either the SMOS product selected in the SNAP, or all SMOS products residing in a specified directory of the file system can be used for exporting. When a directory is specified, the exporting process will convert all SMOS files contained in the directory. The exporter handles unpacked as well as zip-compressed products.
Target Directory: All target files resulting from the exporting process will be stored in the directory specified.
Region-of-Interest: There are three possibilities for specifying a region-of-interest (ROI) considered for export:
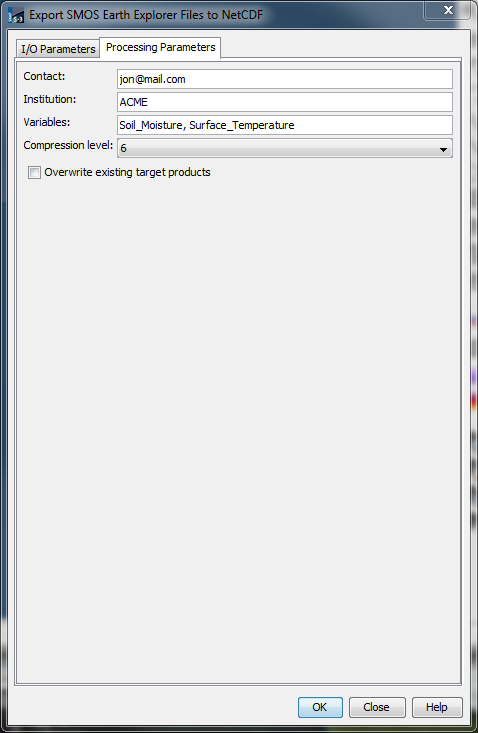
Processing parameters are available in the Processing Parameters tab shown below. Moving the mouse pointer over parameter elements yields further information.
The NetCDF conversion tool is implemented as a GPT operator. This allows to use the converter in batch mode using the GPT command line tool. Information about the Graph Processing Tool can be found in the SNAP main documentation, chapter Graph Processing Framework.
| Name: | NetcdfExport |
| Full name: | org.esa.smos.ee2netcdf.NetcdfExportOp |
| Purpose: | Export SMOS products from Earth Explorer format into NetCDF. |
| Version: | 1.0 |
| Name | Description | Type |
|---|---|---|
sourceProduct |
The source products to be converted. If not given, the parameter 'sourceProductPaths' must be provided. | MIR_BW[LS][DF]1C|MIR_SC[LS][DF]1C|MIR_OSUDP2|MIR_SMUDP2 |
| Name | Data Type | Default | Description | Constraints |
|---|---|---|---|---|
sourceProductPaths |
String[] |
Comma-separated list of file paths specifying the source products. Each path may contain the wildcards '**' (matches recursively any directory),'*' (matches any character sequence in path names) and '?' (matches any single character). | ||
targetDirectory |
File |
. |
The target directory for the converted data. If not existing, directory will be created. | |
geometry |
Geometry |
A geographical region-of-interest as a polygon-geometry in well-known text format (WKT), i.e. POLYGON((...)). | ||
overwriteTarget |
boolean |
false |
Set to 'true' to overwrite already existing target files. |
<S3TBX_HOME>/bin>gpt.bat SmosNetcdfExport <file-path>
<graph id="SMOS test conversion">
<version>1.0</version>
<node id="smos-conversion">
<operator>SmosNetcdfExport</operator>
<parameters>
<targetDirectory>C:/Data/results</targetDirectory>
<geometry>POLYGON((-80 20, -50 20,-50 -20,-80 -20, -80 20))</geometry>
</parameters>
</node>
</graph>
Assuming this XML is written to a file named
roi-example.xml
, the command:
<S3TBX_HOME>/bin>gpt.bat roi-example.xml <file-path>
<S3TBX_HOME>/bin>gpt.bat SmosNetcdfExport "-PtargetDirectory=C:/Data/results" "-Pgeometry=POLYGON((-80 20, -50 20,-50 -20,-80 -20, -80 20))"or using a parameter file:
<S3TBX_HOME>/bin>gpt.bat SmosNetcdfExport -p convparam.txtwhere the file convparam.txt contains key/value pairs of the parameters:
targetDirectory = C:/Data/results
geometry = POLYGON((-80 20, -50 20,-50 -20,-80 -20, -80 20))
<graph id="SMOS test conversion">
<version>1.0</version>
<node id="smos-conversion">
<operator>SmosNetcdfExport</operator>
<parameters>
<sourceProductPaths>C:/Data/SMOS/*SMUDP2*,E:/Data/SMOS/*SMUDP2*</sourceProductPaths>
<targetDirectory>C:/Data/results</targetDirectory>
<geometry>POLYGON((-80 20, -50 20,-50 -20,-80 -20, -80 20))</geometry>
</parameters>
</node>
</graph>
Assuming this XML is written to a file named
directories-example.xml
, the command:
<S3TBX_HOME>/bin>gpt.bat directories-example.xml